#ScienceSaturday posts share exciting scientific developments and educational resources with the KAND community. Each week, Dr. Dominique Lessard and Dr. Dylan Verden of KIF1A.ORG summarize newly published KIF1A-related research and highlight progress in rare disease research and therapeutic development.
KIF1A-Related Research
The architecture of kinesin-3 KLP-6 reveals a multilevel-lockdown mechanism for autoinhibition
For proteins, function follows form. By understanding how different parts of a protein’s structure interact, we can learn more about how mutations and therapeutics impact protein function.
This can present a challenge for KIF1A, which has many flexible domains in its neck whose structure is hard to characterize. These regions can be mutated in KAND but are less understood than motor domain mutations. Research network members like Dr. Arne Gennerich are working hard to improve our knowledge of KIF1A structure directly.
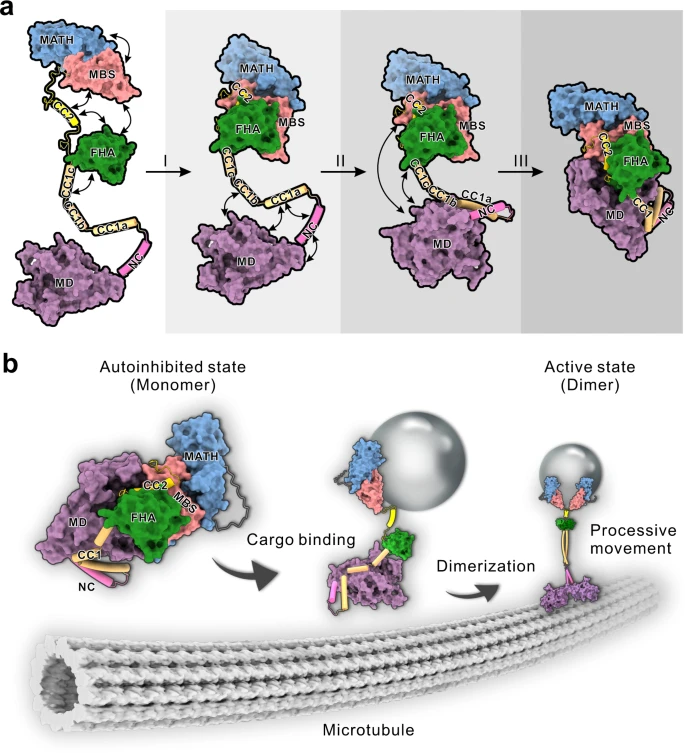
In this week’s article, another team of researchers took an indirect route by characterizing the structure of KLP-6, a kinesin-3 family member expressed in C. elegans. The authors were interested in understanding the neck region of KLP-6 – in most kinesins the neck is thought to participate in autoinhibition, the process of shutting down motor movement until an appropriate cargo has been bound.
The authors found that several domains in the neck cooperate to successively shut down dimer formation, motor activity, and microtubule binding. Disrupting the structure of these inhibitory domains increased KLP-6 movement. The researchers compared KLP-6 and KIF1A structure and hypothesized that mutations in these inhibitory domains could contribute to hyperactive KIF1A, which could have implications for the way KAND manifests in these patients.
KLP-6 is not KIF1A and there are certainly differences in their structure, but investigating these kinds of parallels is crucial for science and medical research to advance.
Rare Roundup
DeepMind’s protein-folding AI cracks biology’s biggest problem
To continue our theme, understanding protein structure is an overarching challenge in research and therapeutic development. Learning about structure can teach us about disease mechanisms and creating effective treatments. But this is a difficult process: Long sequences of amino acids can take on many conformations that can change as they bind with other proteins. In the case of KIF1A, the protein structure of the neck changes a lot depending on whether it is in its inhibited state or is cargo-bound and moving.
The gold standard for understanding protein structure is X-ray crystallography, but this is expensive and time intensive in a way that is difficult to scale up. To accelerate our understanding of protein structures, DeepMind has developed AlphaFold, an AI approach that predicts protein structure based on existing observations of the protein and some basic rules about protein folding. DeepMind is now publishing the predicted structures of over 200 million proteins.
These predictions aren’t perfect: AlphaFold doesn’t always predict structural changes in response to a mutation, for example. But by providing likely structures of so many proteins, this technology has the potential to significantly accelerate molecular research of proteins in health and disease.

