#ScienceSaturday posts share exciting scientific developments and educational resources with the KAND community. Each week, Dr. Dylan Verden of KIF1A.ORG summarizes newly published KIF1A-related research and highlights progress in rare disease research and therapeutic development.
KIF1A-Related Research
Modeling the motion of disease-associated KIF1A heterodimers
This week we’re excited to share a pre-print* from the lab of Research Network member Shinsuke Niwa! His group investigated how we can best predict the movement of KIF1A heterodimers.
What’s a heterodimer? Many people with KAND have a heterozygous mutation: They carry one mutant and one healthy copy of KIF1A. Because KIF1A operates in pairs, these people may have pairs of healthy KIF1A, pairs of mutant KIF1A, or a pair with one healthy and one mutant. This is called a heterodimer (“different pair”). But for many experiments, it is simpler to work with homodimers of a single type of KIF1A. So there’s still a lot we don’t know about how heterodimers of healthy and mutant KIF1A move.
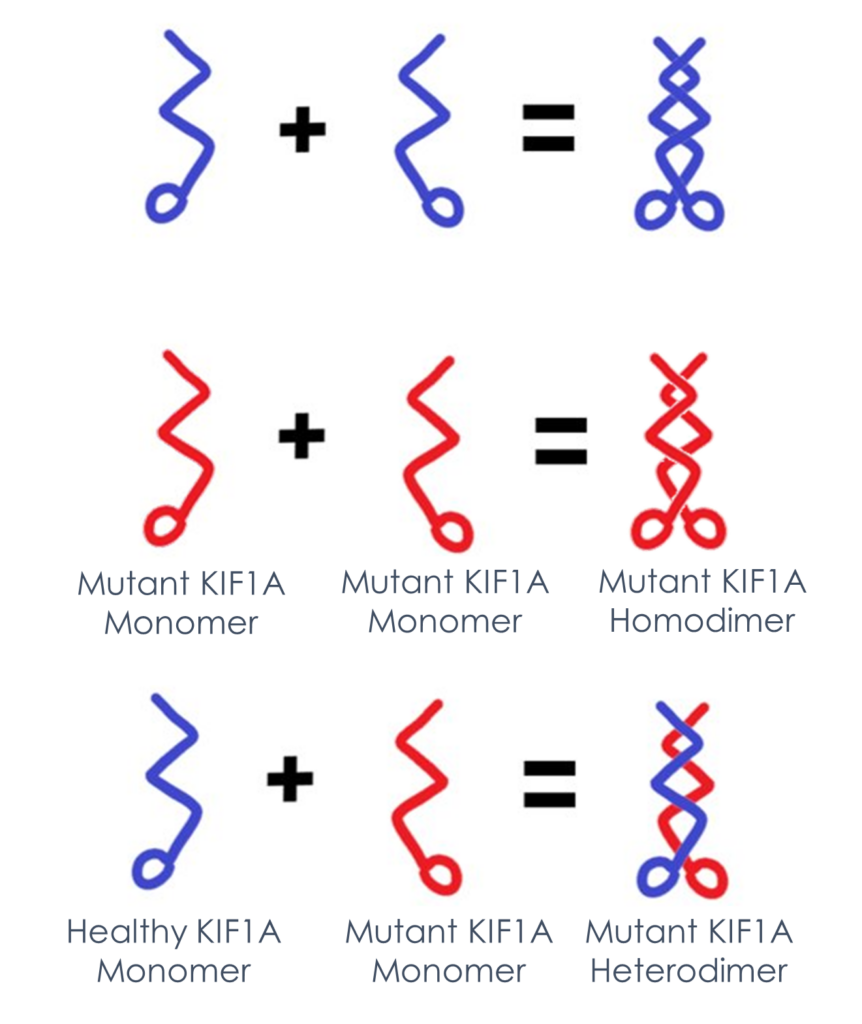
Imagine a 1600m relay race with teams of two, where one runner hands off their baton to the other at 800m. If you had watched each runner perform in a 1600m individual race beforehand, you could roughly predict their relay race time – just average the speed of the two runners. In other words, independent measurements of each runner can be used to predict the results of their relay race.
Using similar logic, researchers measured the movement of KIF1A homodimers (a pair of healthy KIF1A motors, or a pair of mutant KIF1A motors). They then plugged these measurements into equations to predict the movement of KIF1A heterodimers. These independent head models combined measurements of mutant or healthy KIF1A alone to predict how they move together in heterodimers. The accuracy of this model can be used to predict heterodimer properties based off of homodimer experiments, making these studies simpler to conduct.
But this doesn’t account for important factors in the race: handing off the baton might not seem like much, but those few seconds of cooperation between runners could change the results of a race.
For KIF1A, each step involves a relay between its two motors. So we might be able to more precisely predict KIF1A movement if we account for interactions between the two paired motors.
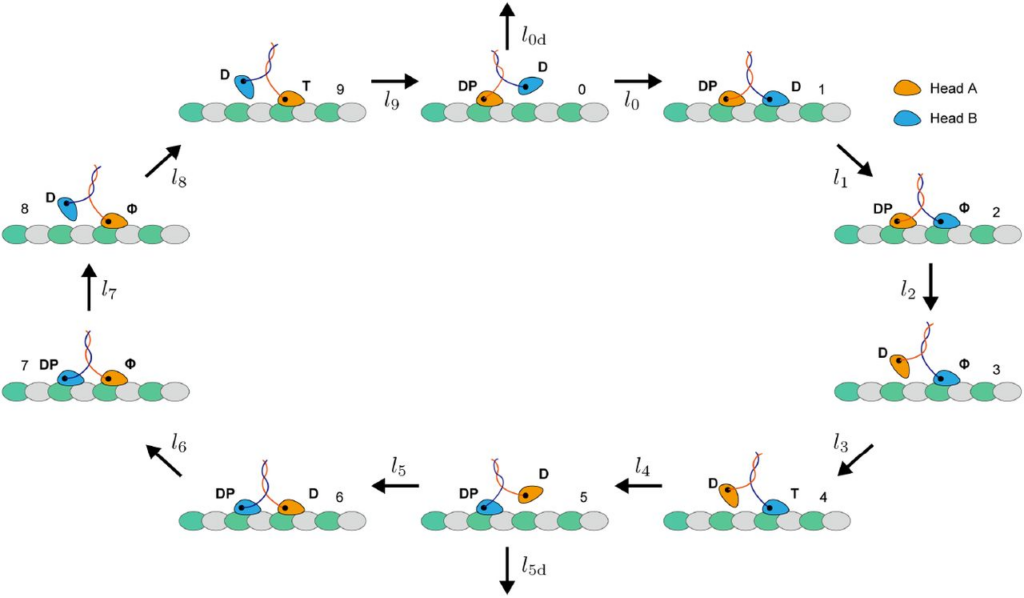
The authors found that as KIF1A walks, its trailing “tethered” head is still actively interacting with the microtubule even though it isn’t firmly planted, which has subtle influences on its movement that can be added to equations for KIF1A movement. This activity may be regulated by the K-loop, which is known to be important for KIF1A’s long-range movement. By interacting with the microtubule, the tethered head may keep KIF1A from detaching mid-stride.
In science, aligning mathematical models with experimental observations improves our understanding of the underlying biology. This study created two models of KIF1A movement that can be used in different types of studies to better predict how .
*What’s a pre-print? Check out this #ScienceSaturday post to learn more.
Rare Roundup
A seemingly small semantic issue is a major roadblock to develop treatments for rare diseases
We’ve spoken often about the goal of repurposing existing drugs to better treat KAND – this is a reality for many families who utilize off-label prescriptions. But drug R&D is an uncertain process, and many therapeutics get shelved during preclinical or clinical research, before they ever reach a patient. But promising drugs that didn’t make the cut for treating one disorder may still be effective for other rare diseases. Taking a drug that shelved during development or didn’t receive FDA approval, and assessing its application for another disease, is called drug repositioning.
Making drug repositioning a part of therapeutic development is a complicated endeavor relying on interactions between regulatory bodies, pharmaceutical companies, rare disease researchers, and patients. It requires a deep look at former products that ended at various stages of research and development. But we may find that we have a wealth of potential small molecule solutions for newly recognized disorders, just waiting to be rediscovered, and repositioned.

