#ScienceSaturday posts share exciting scientific developments and educational resources with the KAND community. Each week, Dr. Dylan Verden of KIF1A.ORG summarizes newly published KIF1A-related research and highlights progress in rare disease research and therapeutic development.
KIF1A-Related Research
Large-Scale Whole-Genome Analysis of HTLV-1–Associated Myelopathy Identified Hereditary Spastic Paraplegias
Many disorders look similar to each other when comparing symptoms; there are many causes of epilepsy, spasticity, and vision loss. But similar looking diseases won’t necessarily respond to the same treatments, which is why it’s so important to find accurate diagnoses.
HTLV-1–Associated Myelopathy (HAM) is a transmissible viral disorder that causes neurodegeneration in the spinal cord, causing weakness and spasticity in the legs. Clinically, HAM appears very similar to hereditary spastic paraplegias (HSPs), which we discussed last week. Because of this overlap, clinicians are interested in finding new criteria to separate the viral HAM and genetic disorders like KAND. In this week’s article, researchers in Japan performed whole genome sequencing on 315 patients who were enrolled in the HAM patient registry.
In addition to sequencing patients, the researchers looked for antibodies to the HTLV-1 virus in the patients’ spinal fluid. Patients with higher levels of antibodies and other signs of inflammation were sorted as having HAM.
Mutations in over 80 genes have been associated with types of spastic paraplegia. 6 patients in the study had mutations in one of these genes, including a single patient with a novel KIF1A mutation, p.V1290F.
The V1290F mutation is a variant of uncertain significance; that means there isn’t enough information about the mutation to know whether it is disease causing. But this patient also had low antibody levels, indicating that HAM was less likely.
Notably, there were many patients in the study who had low antibody levels without a definite genetic cause. This may reflect that there are more unidentified genes that can cause spastic paraplegias, or that the HAM patients were at different stages of disease progression.
While 6 mutations in 315 patients may not seem like much, it provides clarity for patients with genetic HSPs, and helps us get closer to looking for KIF1A mutations in related disease groups.
Rare Roundup
A review of allele-specific antisense oligonucleotides (ASOs) and the handle approach
This week we’ve had many families asking for background info on ASOs, so we’d like to provide a brief overview:
What is an allele?
We each have two DNA copies of the KIF1A gene; each copy is called an allele and makes its own RNA, which is then translated into protein.
Most cases of KAND are caused by a mutation in a single allele of KIF1A. Because KIF1A acts in pairs to walk, the mutant allele can interfere with the healthy allele.
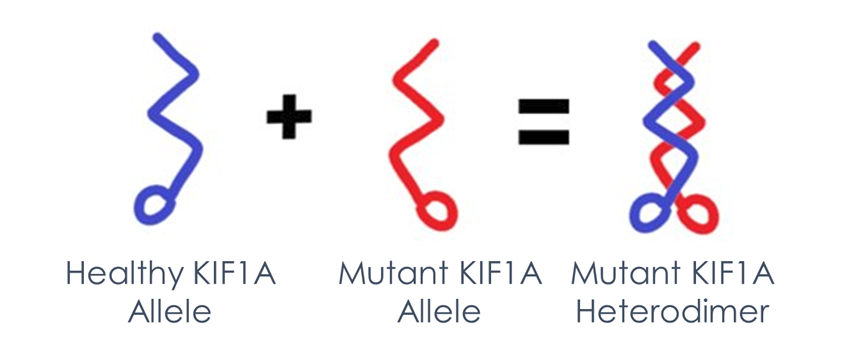
What is an allele specific ASO?
ASO stands for antisense oligonucleotide. It is a type of gene-based therapy for KIF1A mutations that reduces RNA expression of the mutant allele, without interrupting healthy KIF1A expression.
In 2022, n-Lorem worked with the Chung lab to develop an ASO to treat Susannah Rosen, A KAND superhero who has the P305L mutation in KIF1A. This compassionate use n-of-1 trial was the first of its kind for KAND, and the Chung lab has been testing the effectiveness and safety of the ASO for Susannah.
ASOs do not target or change DNA; because they target RNA, which is continuously produced, they are not a one-time treatment. Instead, they must be applied on a regular basis to suppress levels of mutant KIF1A. Because KAND impacts the central nervous system, applying the ASO requires injections into the spinal cord.
Handle approach
ASOs target a specific genetic sequence; traditionally this is the disease-causing mutation, like the one causing Susannah’s P305L mutation. But with over 110 known mutations in our community, developing a single ASO for each individual KAND case would take too much time and resources.
Luckily, not every KIF1A mutation is rare or problematic. Many of us share common mutations called SNPs that don’t cause disease: For example, 3 patients with a P305L, R316W, or R254W mutation may all share a common SNP that is unrelated to their KAND mutation. We can use those SNPs as “handles” for an ASO that treats patients with multiple mutations.
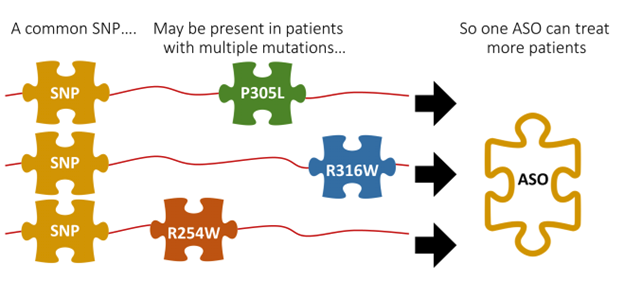
Because SNPs aren’t disease associated, they won’t show up in a standard genetic report. Instead, they must be identified from whole genome sequencing, or whole exome sequencing.
For a patient’s mutation to be knocked down by an allele-specific ASO, they must have the SNP on the same allele that carries the disease-causing mutation. Here are some examples of SNP-mutation mismatches that would not be amenable to ASOs.
- No SNP: Patients without the target SNP would receive no benefit from an ASO.
- SNP on healthy KIF1A: The ASO would knock down healthy KIF1A expression, likely worsening disease course.
- SNP on both alleles: The ASO would cause global KIF1A knockdown, likely worsening disease course.
Over the next month we’ll be discussing ASOs in further detail with our community. In the meantime, please check out our resources on ASOs for more information:

