Want to learn more about KIF1A research without needing a PhD? Through Research Simplified, we collaborate with scientists to create “simplified” summaries of their research and scientific concepts that impact our community. If you have questions about this or other KIF1A-related research, contact us!
KIF1A.ORG thanks Dr. Kaur and Prof Christodoulou for summarizing their work for us, and all they do to advance KIF1A research and therapeutic development. Learn more about our collaboration with the Christodoulou Lab on our Funded Projects page.
Expansion of the phenotypic spectrum of de novo missense variants in kinesin family member 1A (KIF1A)
Authors: Simranpreet Kaur, Nicole J. Van Bergen, Kristen J. Verhey, Cameron J. Nowell, Breane Budaitis, Yang Yue, Carolyn Ellaway, Nicola Brunetti‐Pierri, Gerarda Cappuccio, Irene Bruno, Lia Boyle, Vincenzo Nigro, Annalaura Torella, Tony Roscioli, Mark J. Cowley, Sean Massey, Rhea Sonawane, Matthew D. Burton, Bitten Schonewolf‐Greulich, Zeynep Tümer, Wendy K. Chung, Wendy A. Gold and John Christodoulou
Research Simplified By: Dr. Simranpreet Kaur and Prof John Christodoulou
Dr. Simranpreet Kaur
Simran is a young early career researcher who has more than 12 years’ of professional experience in genetics and molecular biology with a committed track record in the field of rare disease in children. Untangling the underlying genetic and molecular causes of the problems in affected children is of particular interest to her. Her research vision is to connect, collaborate and contribute towards the scientific journey for finding the reason and solutions in children facing challenging health problems, thus shortening their diagnostic odyssey and improving their overall quality of life.
Prof John Christodoulou
John has over 25 years’ experience as a paediatric geneticist clinician scientist. He has always been interested in understanding how genetic disorders can affect brain function, and his research interests have focused on this issue. He is best known for his clinical and laboratory research into Rett syndrome, a devastating neurodevelopmental disorder predominantly affecting females, for which there is currently no effective treatment. He has co-authored over 270 research papers (over 80 relating to Rett syndrome and related disorders), 11 major reviews and contributed to 15 book chapters.
Communication In Our Nervous System
The human brain is a very complex organ made up of billions of cells called neurons. Although not physically connected, neurons communicate with each other effectively through specialized connection points called synapses to synchronize brain function. Structurally, the neuron consists of dendrites, a cell body, axon and neurite terminals (Figure 1). Most of the proteins critical for healthy synapse function are made in the cell body of the neuron, which are then packaged as ‘cargo’ and transported along the axon towards the neurite terminals in a rapid and well coordinated manner.
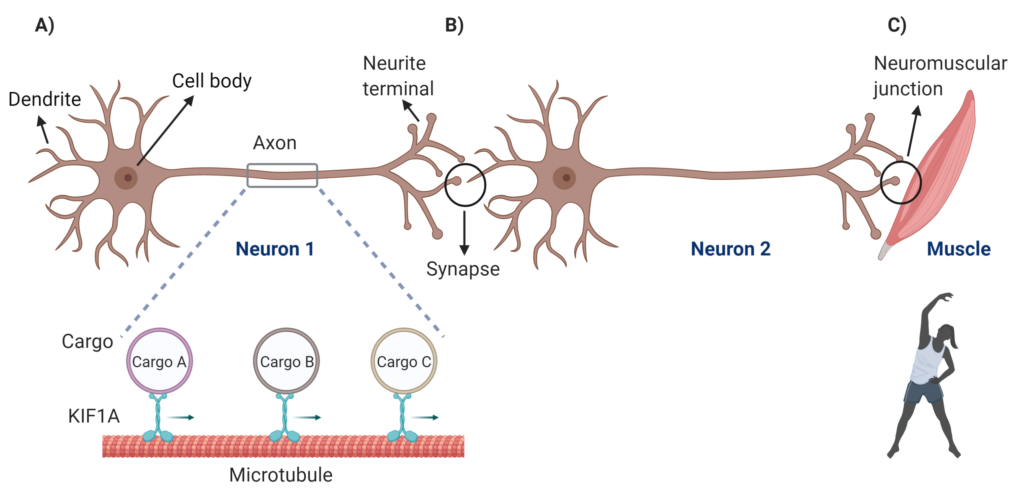
Figure 1: Schematic showing neuron structure, communication and muscle control. A) Part of an axon is magnified to show KIF1A transporting cargo along microtubule network from the cell body towards neurite terminals B) Neurons communicating through the connection point called a synapse C) the neuromuscular junction, a specialized region through which the message is passed from the neuron to the muscle to activate it to contract. Created with BioRender.com
KIF1A and the Importance of Neuronal Cargo Delivery
Any small hold-up in the delivery of critical cargo can disturb the function of neurons, potentially leading to progressive irreversible degeneration (death) of neurons, creating havoc in the brain. The kinesin superfamily proteins (KIFs) are ‘trucks’ that efficiently carry critical cargo to their required locations, by moving along the microtubule highways using their ‘engines’, the motor domains, which generate the power to move them by using ATP as the ‘fuel source’. The Kinesin family member 1A gene (KIF1A) encodes for the motor protein KIF1A, which is specifically found in neurons, and is involved in the fast delivery of its specific cargo to the tips of neurons, supporting healthy brain function. In affected individuals, faulty trafficking by dysfunctional KIF1A due to a change (variant) in the KIF1A gene causes delayed delivery of its cargo in neurons, triggering a devastating spectrum of rare brain disorders collectively referred to as KIF1A-Associated Neurological Disorder (KAND), which is known to affect approximately 300 people, mostly children, worldwide. KAND symptoms often appear at birth or early childhood, have variable severity, and sadly may result in death within 5 years of life. Because clinical features overlap with other neurological disorders, a significant number of people remain either misdiagnosed or undiagnosed, and so we think that the figure of 300 children worldwide is a considerable underestimate.
KAND and Rett Syndrome: How Do They Overlap?
Our lab explores the hidden genetic aspects of various neurological disorders including Rett syndrome (RTT), a devastating paediatric-onset severe rare neurodevelopmental disorder. Although RTT is mainly caused by variants in the methyl-CpG-binding (MECP2) gene, up to 15% of RTT individuals do not have a genetic diagnosis. We used various advanced genetic techniques to end the diagnostic odyssey for affected families by uncovering the gene responsible for the clinical symptoms. During our recent study, we identified a variant in the motor domain of KIF1A in one individual with a clinical diagnosis of RTT. Through international collaborations we identified three additional individuals with clinical features overlapping with RTT who we found to have genetic variants in KIF1A.
How Does The Christodoulou Lab Study KAND?
Various computer based tools predicted that the identified variants were likely to have damaging effects on the function of KIF1A in the cell. KIF1A is almost exclusively expressed in neurons, thus creating a major practical challenge to study this critical protein directly in neuronal cells from an individual’s brain. We therefore developed a number of cell and molecular biology-based resources to study normal and dysfunctional KIF1A using readily available human brain-like (SH-SY5Y) cells that are commonly used to study brain-related proteins. To see KIF1A under a microscope, we were able to manufacture KIF1A proteins with a specialized tool that shines green under a special type of light. This allowed us to track movement of the KIF1A protein within live cells in “real time”. While the normal KIF1A rapidly moved through the cells and accumulated at the neurite tips, KIF1A with KAND individual specific variants remained localized near the cell body, suggesting that the mutated KIF1A failed to move effectively along the microtubule network (Figure 2).
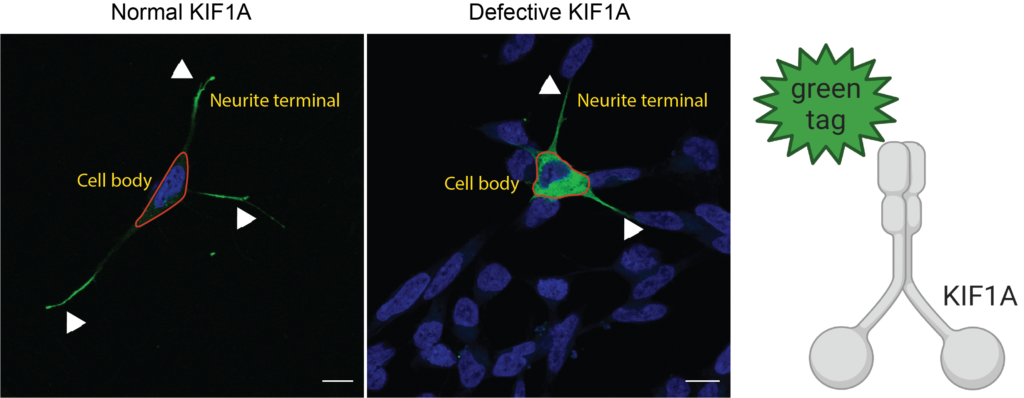
Figure 2: Human brain-like cells (SH-SY5Y) showing accumulation of normal KIF1A (green) at neurite tips and KIF1A with KAND variant stuck near the cell body (Blue = nucleus). Created with BioRender.com
We also developed another “test in a dish”, the microtubule gliding assay (Figure 3). In this assay, we created a “lawn” of of either normal or mutated KIF1A, and then apply synthetic microtubule fragments that are tagged with a special tag that emits red color when looked under microscope, and capture movement of the microtubule fragments along the KIF1A lawn under a microscope.
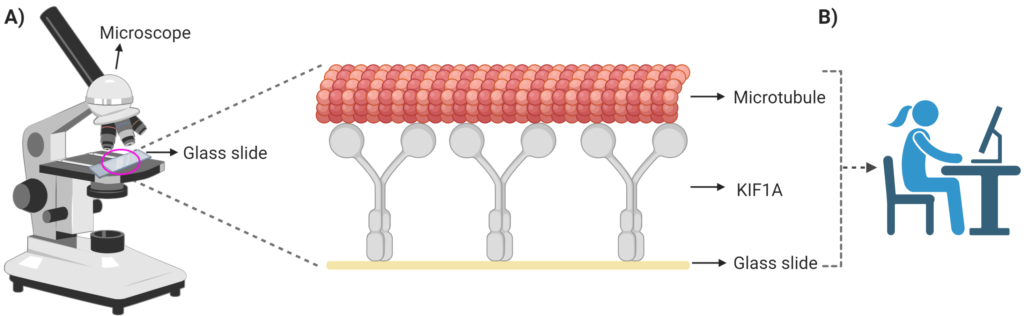
Figure 3: Schematic showing microtubule gliding assay. A) Set up of the assay with a zoom into the glass slide showing the “lawn” of the KIF1A protein upon which the microtubules move. When exposed to a special light the microtubules emit a red colour which allows us to track their movement under the microscope. B) Computer based methods are used to calculate the speed with which the microtubules move over the normal KIF1A or KIF1A with KAND variants. Created with BioRender.com
The videos showed that most of the microtubule fragments were able to glide along the normal KIF1A lawn in a snake-like pattern (Video 1).
However for the mutated KIF1A lawn, microtubules were either not able to attach or moved much more slowly (Video 2, Video 3 and Video 4).
What Can We Learn From This Study?
From these studies we conclude that:
- KIF1A variants can cause a broad range of clinical pictures, including RTT.
- The molecular assays we have developed to study KIF1A in collaboration with Prof Kristen J. Verhey and her team at the University of Michigan are very powerful tools for understanding the effect of KIF1A mutations.
- This work has opened up avenues to explore personalised therapies for KAND individuals that we hope will have a significant impact on affected children and their families.
Do you have any questions about what you just read? Contact us to share your questions and feedback!

This is an amazing assay to have available for research. Is it expensive to produce? Is it able to be modified by mutation variant? I am asking because it would be interesting to try to score phenotypic severity relative to microtubule motion on this assay. I wonder if small molecule experiments on this assay are possible. Very cool!
[…] Read the Research Simplified […]