#ScienceSaturday posts share exciting scientific developments and educational resources with the KAND community. Each week, Dr. Dominique Lessard and Dr. Dylan Verden of KIF1A.ORG summarize newly published KIF1A-related research and highlight progress in rare disease research and therapeutic development.
KIF1A-Related Research
Optogenetic control of kinesins and dynein reveals their specific roles in vesicular transport
KIF1A is only one of over 40 kinesin motor proteins in mammals, so why do KIF1A mutations have such drastic effects?
Well, each of these kinesins is specialized – they may be expressed in specific cell types or transport different cargo. But in many cases, multiple types of kinesins are bound to the same cargo; this raises the question of how each type of kinesin is contributing to transport.
In this week’s article, researchers used a technique called optogenetics to look at how different motor proteins collaborate – and compete – to move cargo.
Optogenetics
One of the most effective ways to see what a protein does is to turn it off and see how the system behaves in the absence of that protein; traditionally this would be done by “knocking out” the gene for that protein, preventing it from being made at all.
This approach can teach us a lot about a system, but it’s a bit like being handed a toy car’s remote control without batteries – you can see that the car isn’t moving, but you don’t know how the controls are supposed to work, or how fast it’s supposed to move.
But what if you were given a controller that had charged batteries and an on/off switch? You could see what happens when you turn off the controller when the car’s moving at full speed, or taking a turn. This can give you much richer moment-to-moment information about how the car works.Opto-(“light”)-genetics works on a similar principle. The first optogenetic proteins were derived from bacteria, and changed their structure in response to light, much like an on/off switch – by adding these switches to other proteins, researchers can turn proteins on and off with light, manipulating them in real time. You can learn more about the principles of optogenetics in the video below:
The Study:
The authors were investigating in the interactions between four families of motor proteins:
- Kinesin-1, Kinesin-2, and Kinesin-3 (KIF1A’s family): These motors have a similar structure with key differences that help them work with specific cargo, walk along different microtubules, and walk in specific patterns.
- Dynein: This motor moves cargo toward the cell body, in opposition to kinesins.
All of these motors interact with a common cargo called an endosome: These are like pallets that hold several types of cellular materials going to the same place; depending on the materials, the endosome may have different destinations.
As endosomes are sorted they mature from early endosomes to late endosomes that have different surface markers and contents. Late endosomes then merge into acidic lysosomes that break down cellular waste.
To study how different motor proteins contribute to endosome trafficking, the authors made optogenetic switches to turn off each of these motors one at a time.
When all motors were active, the endosome spent a lot of time moving forward and backward, representing a tug-of-war between multiple motors that helped sort the endosome.
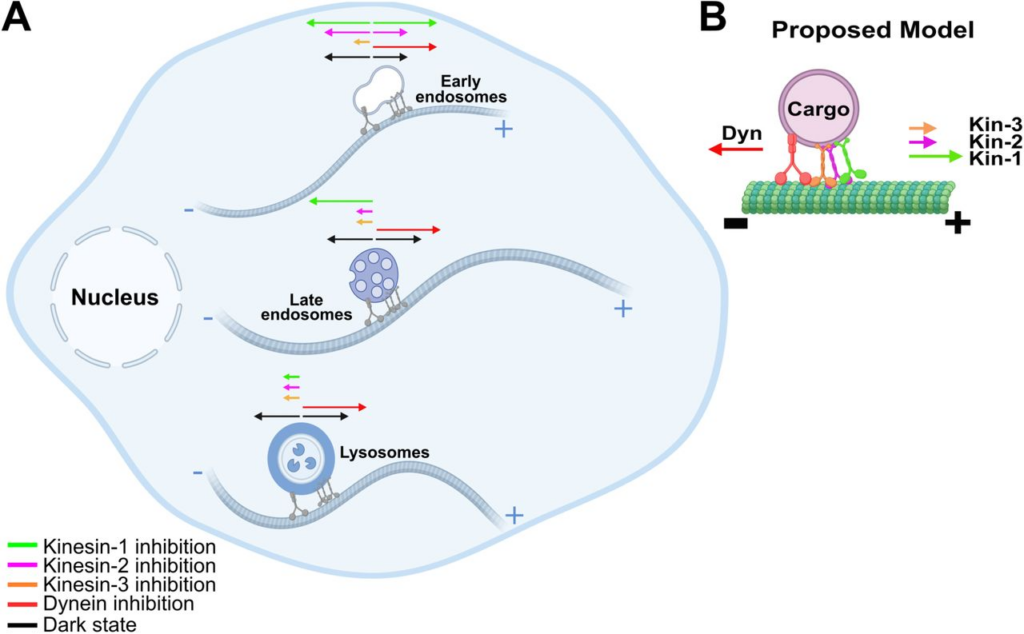
Turning off kinesin-1 and dynein actually made early endosomes move faster, with less stops and reversals; these motors appear to participate in a tug-of-war that slows down cargo to steer it to the right destination.
On the other hand, inhibiting kinesin-3 (KIF1A) caused early endosomes, late endosomes, and lysosomes to stall or reverse direction. This supports its role as a persistent, long-haul motor throughout endosome transport.
Turning off kinesin-2 also stalled or reversed late endosomes and lysosomes, but didn’t impact early endosomes undergoing sorting. Kinesin-2 and kinesin-3 may overlap in some stages of long-range transport, but kinesin-3 could be particularly important for early endosomes when kinesin-2 isn’t engaged.
Science itself is a tug-of-war between simplifying systems to ask specific questions, and acknowledging the complexity of the system so we can get the whole picture. It’s important to remember that KIF1A is just one piece of a large network that sorts and traffics cells/cellular components; deciphering how it works with a motor protein team can help us learn how KIF1A mutations impact neuronal health.
Rare Roundup
FindZebra Enhances Rare Disease Information Retrieval through Integration with OpenAI’s GPT API
When it comes to diagnosing diseases, there’s an old saying in medicine: “When you hear hoofbeats behind you, don’t expect to see a zebra.” In other words, don’t look for an exotic or rare disease when a more common one is likely.
This warning has its merits; when you have limited time and resources, it’s efficient to look for common diseases first. But with hundreds of millions of individuals in the world affected by one of 7,000-10,000 rare diseases, zebras are more common than ever. The trick is developing tools that make it more efficient to recognize them, and keeping those tools relevant as our knowledge and technology advances.
This is the objective of FindZebra, which created an online search engine that provides potential rare disease associations based on symptoms or genes. FindZebra can’t diagnose, but it can make it easier for physicians to identify potential diseases, ideally reducing misdiagnoses.
FindZebra recently updated its search by integrating OpenAI’s GPT into its algorithm after training it on medical board exam questions. As the field of language modeling progresses, the hope is that this update will add nuance to search results and identify rare disease hoofbeats more accurately.

