Want to learn more about KIF1A research without needing a PhD? Through Research Simplified, we collaborate with scientists to create “simplified” summaries of their research and scientific concepts that impact our community. If you have questions about this or other KIF1A-related research, contact us!
A Highly Conserved 310-Helix Within the Kinesin Motor Domain is Critical for Kinesin Function and Human Health
Authors: Aileen J. Lam, Lu Rao, Yuzu Anazawa, Kyoko Okada, Kyoko Chiba, Mariah Dacy, Shinsuke Niwa, Arne Gennerich, Dan W. Nowakowski, Richard J. McKenney
Research Simplified By: Aileen Lam and Dr. Richard J. McKenney
Aileen Lam
Aileen is a former member of the McKenney Lab at the University of California – Davis and helped conduct this research when she was a lab technician. Aileen served as the Science Communication Associate at KIF1A.ORG from November 2020 – July 2021. Now, she’s off to med school at Western University of Health Sciences where she will continue to combine her passions for research and patient advocacy to serve those who are underrepresented in medicine.
Dr. Richard J. McKenney
Richard earned his Ph.D. at Columbia University where he studied the biophysical properties of the cytoplasmic dynein motor with Professor Richard B. Vallee. Richard then moved to the laboratory of Dr. Ronald Vale, where he continued to research the mechanism and regulation of the dynein motor. The McKenney lab at the University of California – Davis maintains a strong interest in the biochemical and biophysical mechanisms of the molecular motor proteins dynein and kinesin, and how these mechanisms relate to human diseases.
KIF1A, the Critical Cargo Transporter
The human body is extremely complex, with many moving parts that are still under investigation to this day. On a molecular level, the cells that make up our body require many different types of molecules for various functions, and these components must often be transported to their proper locations within cells. This process of cellular transport is absolutely essential to our health, function, and development, as it is responsible for delivering important cargoes throughout our bodies. To facilitate cellular transport, KIF1A, a motor from the kinesin family of proteins, plays a key role in actively moving cellular cargo within neurons along tracks called microtubules. This process is critical for healthy brain formation and function. Mutations in the KIF1A gene result in defects in the motor’s ability to properly carry out its transport function, leading to a rare neurodegenerative disease known as KIF1A-Associated Neurological Disorder (KAND). Currently, there are over 100 known mutations reported that cause KAND in approximately 300 people and, as of today, there is no cure for this disease, making the urgency to spread awareness and conduct research crucial for the rapid development of a treatment.
What Makes KIF1A Unique?
There are over 40 different kinesin genes in humans that code for kinesin motor proteins and KIF1A is one of several motors that belong to the kinesin-3 subfamily. Kinesin-3 motors are characterized by their very high microtubule binding rate (aka a strong likelihood for KIF1A to land and attach to microtubules) and its ability to travel further and faster along microtubules compared to other kinesin family groups. This special feature of KIF1A is an area of active research. It is thought that a specific region within the KIF1A motor called the K-loop, which contains positively charged amino acids, plays an important role. With microtubules acting as negatively charged “tracks” that KIF1A motors travel on, the positively charged K-loop is thought to be responsible for KIF1A’s strong interaction with microtubules. Additionally, as all proteins adopt specific 3-dimensional structures, or shapes, that are unique and required for a protein’s specific function, being able to uncover the structure of the K-loop would bring more insight to its role in the KIF1A motor. As of today, scientists have determined most of KIF1A’s 3-dimensional structure, but the K-loop structure remains unknown, necessitating further research into how the K-loop exerts its effects on KIF1A’s ability to transport cargo.
What We’ve Learned: The Mystery of the K-loop and its Relation to KAND
Working with KIF1A.ORG, we became aware of a novel KIF1A mutation, P305L, that is located right next to the K-loop. This P305L notation means that the 305th amino acid, which in this case is a proline, was mutated into a leucine. The close proximity of the P305 amino acid to the K-loop suggests that it could play a key role in regulating the activity of this critical region. Additionally, P305 is part of a highly conserved amino acid sequence (PYRD) that is present in most other kinesin family proteins. In this work, this sequence is found to adopt an unusual 310-helical shape, which is characterized as a shorter more tightly wound helix when compared to the much more common α-helices that make up a majority of known protein structures (Figure 1). Furthermore, proline residues are recognized for their ability to mediate shape changes in proteins due to their ring framework, which suggests that this amino acid could play a role in the structure of the 310-helix (Figure 2).
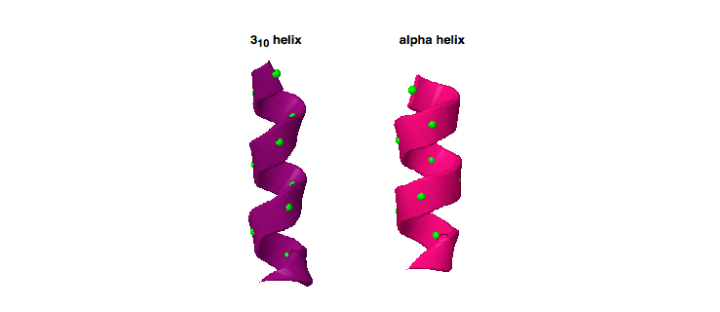
The image here is of two helices, which are structures commonly found to make up proteins and are composed of amino acids (green balls) bonded together to create a helical shape. This comparison highlights the differences seen between an α-helix (right, pink) and a 310-helix (left, purple). When looking at the helices, notice how the 310-helix is more tightly wound than the α-helix. The difference in this feature can give proteins different dynamics and/or functional outcomes (PDB ID 3L79_514-525 and PDB ID 1HHO_B_5-16).
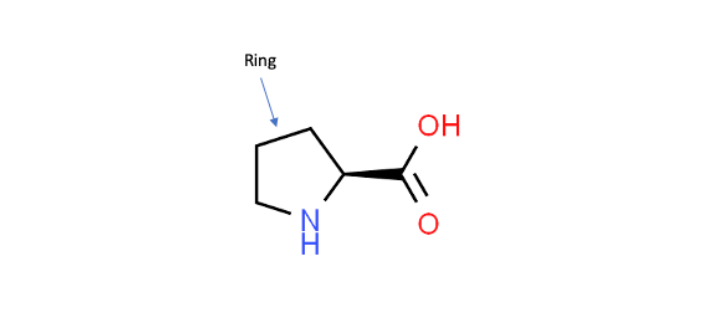
Shown here is the amino acid proline with its unique ring structure. Because of its framework, proline can play a role in affecting a protein’s shape (Modified from ChemSpider ID128566).
Recognizing that the 310-helix may be essential to the proper function of KIF1A, we conducted a comprehensive analysis of the P305L variant using biochemical and biophysical, genetic, and single-molecule approaches. By comparing wild-type and mutant P305L KIF1A proteins, we found that the P305L variant reduces KIF1A’s speed, ability to generate force, and time spent traveling on microtubules. Most dramatically, and consistent with the idea that the mutation may impact the function of the K-loop, the P305L variant decreased the rate at which KIF1A motors were able to land on microtubules by over 80%. These results show that although the K-loop facilitates a high microtubule-association rate, the presence of that structure alone is not sufficient for this function, suggesting that a specific functional conformation, or orientation, of the K-loop is mediated by the adjacent PYRD sequence. We propose that the 310-helix, which is composed of the PYRD sequence, is critical for facilitating a specific conformation of the K-loop that is essential for KIF1A function (Figure 3).
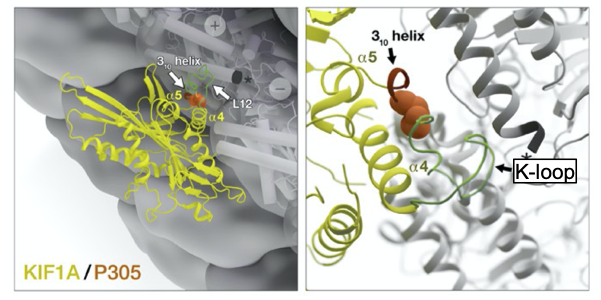
Figure 3. Visualization of 310-helix, P305, and K-loop
In these computer-generated model images, KIF1A (yellow) is shown in detail to highlight the locations of P305 (orange), the 310-helix (red loop), and the K-loop (green). Working together, these elements help to carry out KIF1A’s function (modified from Aileen J. Lam et al. Sci Adv 2021;7:eabf1002).
Additionally, by changing the analogous amino acid in KIF5B, a member of a different kinesin subfamily, we observed a similarly dramatic decrease in the kinesin-microtubule interaction, suggesting a common mechanism among the entire kinesin family. The high conservation of this PYRD sequence amongst other kinesin family members alludes to its critical role in motor function and human health. The data also suggest that the KAND phenotype observed in patients harboring P305L mutations, and possibly other mutations in the PYRD sequence (Y306 and R307), results from KIF1A motors that cannot effectively interact with their microtubule tracks. This finding can potentially give more insight into understanding related variants that also fall within the PYRD sequence, as they would be hypothesized to behave similarly to the P305L variant by affecting the microtubule-association rate in a comparable manner.
Overall, the data contribute to our understanding of the structural elements that play a critical role in KIF1A’s motor function. Recognizing the significance of the 310-helix in relation to the K-loop demonstrates that the conformation of the K-loop, not its existence alone, is essential for KIF1A’s proper microtubule association. The conclusions drawn from this paper propose that therapies seeking to enhance KIF1A’s ability to interact with microtubules could possibly be most effective in these particular cases. On a broader scale, considering that the PYRD sequence is highly conserved amongst a range of kinesins, these findings can help inform us of other kinesins and their shared structural properties.